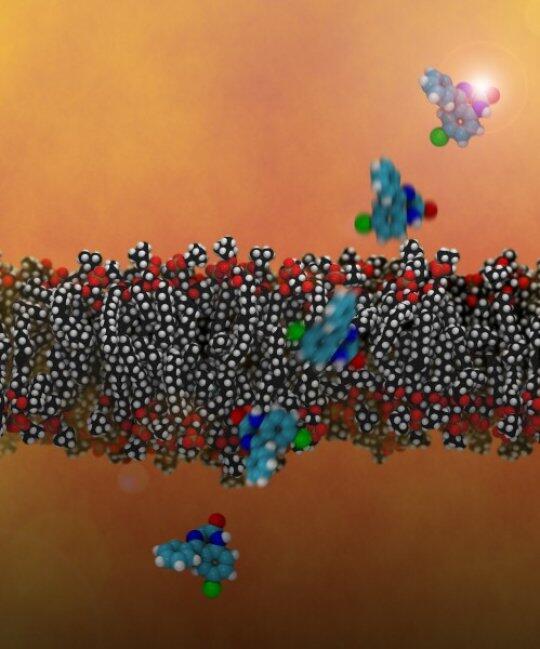
Imagine you wanted to know how much energy it took to bike up a mountain, but couldn’t finish the ride to the peak yourself. So, to get the total energy required, you and a team of friends strap energy meters to your bikes and ride the route in a relay, then add up your individual energy inputs. Researchers at Lawrence Livermore National Laboratory in Livermore, Calif., are currently using a similar approach, powered by LLNL’s world-class supercomputers, to simulate the energy requirements for candidate drug molecules to permeate cell membranes — shaving weeks of compound testing by determining in advance how readily they’ll enter cells to perform their activity.
“Instead of having one [drug molecule] starting from one side of the membrane, you have it starting at a hundred different points through the membrane,” said Timothy Carpenter, a staff scientist in LLNL’s Biochemical and Biophysical Systems Group.
At each of these points, the simulation imposes an artificial force of varying degree on the molecule to keep it in place. By measuring the degree of fluctuations and movement of the molecules at each of these positions, the program can obtain the related energy levels, which can then be stitched together to generate a progressive energy profile. From here, the researchers can also calculate the diffusion rate of the compound — generally determined by the size of the molecule — which they combine with the energy profile to get the permeability rate. (For more information, see the group’s earlier paper, “A method to predict blood-brain barrier permeability of drug-like compounds using molecular dynamics simulations” link below.)
The alternative — running a single simulation for the duration it would take the compound to get through the membrane — is extremely time consuming, as the chances of the compound getting through with a high energy barrier are significantly lower.
Carpenter and his colleagues will present their work this week at the Biophysical Society’s 60th Annual Meeting in Los Angeles, Calif. The group, which also includes LLNL biomedical scientist Nicholas Be and organic chemist Carlos Valdez, is currently focused on developing more permeable oxime-based compounds that can act as treatments for nerve agents, such as sarin gas. These compounds — which are classified by the United Nations as weapons of mass destruction — act by interrupting the breakdown of the neurotransmitter acetylcholine by the enzyme acetylcholinesterase. This results in profuse salivation, convulsions, involuntary bodily eliminations and eventual death by asphyxiation.
Current treatments for nerve agents consist of concurrently administered oxime compounds — typically pralidoxime and atropine — which act, respectively, by re-cleaving the affected acetylcholinesterase and blocking the overstimulation of the acetylcholine receptors by accumulation of the neurotransmitter, concurrently restoring function while treating symptoms. However, the currently used oximes suffer from poor blood-brain barrier penetration.
Those compounds are very effective at reversing the effects of a nerve agent once they get in the brain, but the brain itself is very poorly permeable, and it’s hard to get the drugs across the blood-brain barrier. “The goal of the project was to see if we could develop or improve on the permeability of some of these classes of compounds,” Carpenter said. “It raises the question of: if you have a drug that is half as effective but four times as permeable, is that a better option than what we have at the moment?”
According to Carpenter, it takes about six weeks from start to finish — proposing a drug candidate, getting the ingredients shipped, synthesizing it, and performing in-vitro testing — to assess a drug’s membrane permeability. While this process is still essential for determining a drug’s potential, the simulations allow the researchers to assess a candidate’s membrane permeability in a matter of 16 hours — allowing them to effectively pre-screen drug compounds before synthesizing them, saving over five and a half weeks of effort.
The simulations are run on LLNL’s supercomputers, two of which — Vulcan and Sequoia — are currently both ranked in the top 20 of the fastest supercomputers in the world. Those computers, which are both IBM Blue Gene/Q systems, are respectively capable of five and twenty petaflops, or five and twenty thousand trillion floating operations per second. Each set of 100 simulations takes about 100,000 computer hours — the equivalent of three years on a single quad-core laptop.
Carpenter and his colleagues, including Brian Bennion, Mike Malfatti, Heather Enright, Victoria Lao, and Felice Lightstone at LLNL, Windy McNerney at the War Related Illness and Injury Study Center, and Emma Carlson at the U.S. Naval Academy, plan to continue researching more permeable anti-nerve agents, as well as implementing the methods in other drug design programs.
Source: Science Daily